Rhizobium Bacteria
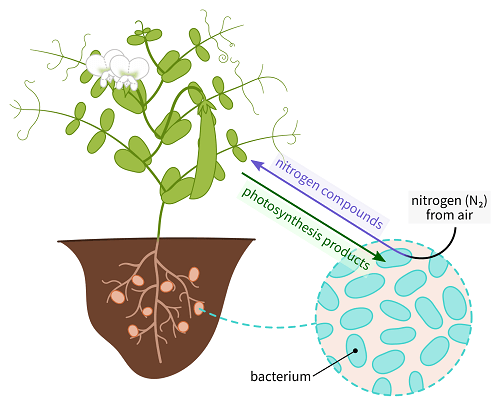
Rhizobia refers to the Rhizobium genus members. However, people’s perspective has changed with time, and now any bacteria that can fix nitrogen and nodulate with legumes come under the genus rhizobium.
History
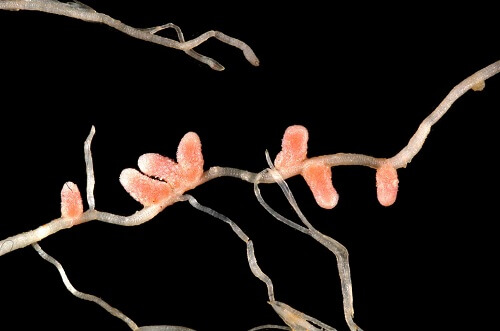
At the end of the nineteenth century, it was discovered that legume plants’ root nodules were digesting atmospheric nitrogen. Beijerinck isolated the root-nodule bacteria in 1888 and proved that they were responsible for the nitrogen fixation process. Bacillus radicicola was the name he gave to these germs. Frank later altered the name to Rhizobium, which had just one species at the time, R. leguminosarum.
In the early 20th century, various researches were done to check the diversity of bacteria that could nodulate the legume host. As a result, cross inoculation groups were formed, which comprised rhizobia from one plant species that could nodulate other plant species belonging to the group.
In the early twentieth century, extensive research of different bacteria’s ability to nodulate various legume hosts led to cross-inoculation groups, with rhizobia from one plant in the group thought to nodulate all other plants in the group. This idea was also utilized in rhizobial taxonomy, but it was eventually abandoned as an unreliable taxonomic identifier, owing to aberrant cross-infection within plant groups. In the early 1960s, bacteriologists began using a wide range of morphological, nutritional, and metabolic characters and serology and simple DNA characteristics in numerical taxonomy studies. This established the close relationship between Rhizobium and Agrobacterium and clearly differentiated between rapid and slow-growing rhizobia, with the latter genus Bradyrhizobium being created. Additional variety was identified among the rhizobia, and their interactions with other groups of bacteria became evident from the 1980s onwards, as more genetic traits (DNA-DNA, and DNA-rRNA hybridizations, rRNA catalogs, rDNA sequencing) were introduced. As a result, the number of genera has gradually increased. The number of legitimately reported species has increased significantly, with 48 rhizobia species currently recognized.
The following are two major explanations for the growth in the number of genera and species:
(1) Numerous legume species have now been investigated. This is in contrast to earlier attempts, which focused on significant food and pasture species crops, largely from the Western hemisphere.
There are a total of 18000 species, out of which only 20%, that is, 650 genera of legume plants, have been investigated for nodulation. This leaves a vast number of legume species to be examined and the possibility of many more rhizobia species and genera to be described.
(2) The continual development of the taxonomic study is another cause for the growing number of rhizobial species. Improvements and breakthroughs in technologies for studying cell DNA and RNA have resulted in more thorough characterization and phylogenetic and polyphasic classifications. A growing number of complete bacterial genomes are becoming accessible at this time. This will very certainly have a significant influence on bacterial taxonomy. Most recent taxonomy investigations have used a polyphasic approach, combining genetic, phenotypic, chemotaxonomic, and phylogenetic data to fully understand the bacteria’s connections and suggest an appropriate categorization.
Sinorhizobium
Different scientists suggested a new genus, Sinorhizobium fredii, for the fast-growing soybean rhizobia, renaming R. fredii as Sinorhizobium fredii and proposing a new species, S. xinjiangense. This new genus sparked debate at first since there was no genetic evidence to support its formation and distinguish it from R. fredii at the time. Later, phylogenetic evidence was offered to establish a third rhizobia genus that was not limited to fast-growing soybean rhizobia, and the genus definition was revised. S. meliloti was suggested for isolates from Acacia and Sesbania from Senegal. Two further species, S. saheli and S. terangae (the initial spelling S. teranga was subsequently changed) were proposed for isolates from Acacia and Sesbania from Senegal. Sinorhizobium is now largely recognized as a genus with 11 genuine species. New genetic evidence has been offered to suggest the split of S. xinjiangense and S. fredii. However, this is based on DNA-DNA hybridizations using DNA from the S. fredii type strain USDA 205T, the quality of which has not been confirmed by homologous hybridization with S. fredii strains. Ensifer adhaerens is phylogenetically a part of the Sinorhizobium lineage, as shown by 16S rDNA comparisons lately.

This organism is a soil bacterium that can cling to and lyse other soil bacteria, and it was first characterised mostly based on phenotypic information. Based on DNA-DNA hybridizations and phenotypic traits, our polyphasic experiments have indicated that a small group of four different rhizobial isolates and two soil isolates cannot be discriminated clearly from Ensifer adhaerens, and we should therefore include these rhizobia in Ensifer. In the 16S rDNA dendrogram of the alpha-Proteobacteria, Ensifer and Sinorhizobium constitute a single group and may be considered a single genus. Because the earlier name Ensifer would prioritize, this has significant nomenclatural implications. There are various reasons why switching from Sinorhizobium to Ensifer would not be the optimal answer and why permitting an exemption to Rule 38 could be preferable. As a result, it was suggested the name Sinorhizobium adhaerens comb. nov. and made a request for an opinion on Sinorhizobium adhaerens’ conservation over Ensifer adhaerens. People thought this idea was unreasonable and advocated that all Sinorhizobium species be relocated to Ensifer instead. While the request for an opinion is being processed, the term Sinorhizobium adhaerens is no longer valid, and Ensifer adhaerens is the right name.
Mesorhizobium
Mesorhizobium is a phylogenetic grouping of five rhizobial species (R. loti, R. huakuii, R. ciceri, R. Mediterranean, and R. tianshanense) that are phylogenetically similar but different from Rhizobium, Agrobacterium, and Sinorhizobium. They have a growth rate that is halfway between that of fast-growing and slow-growing rhizobia. Mesorhizobium is phylogenetically separated from fast-growing rhizobia by Bartonella, Defluvibacter, Aquamicrobium Phyllobacterium, Aminobacter, and Pseudaminobacter, according to 16S rDNA sequence data.
Bradyrhizobium
Rhizobium japonicum, a slow-growing species, was given the name Bradyrhizobium. Although it was established that slow-growing strains exist on many legume genera, the soybean-nodulating B. japonicum was initially the sole species reported. Five other species have been validly named in this genus: B. elkanii and B. liaoningense, B. yuanmingensenodulating Lespedeza, B. betae from tumor-like deformations in the roots of Beta vulgaris.

A variety of serogroups have been identified among slow-growing soybean symbionts in addition to the species division. Bradyrhizobium sp., followed by the name of the legume host, has been isolated from several different slow-growing rhizobia from other legume hosts. Some Bradyrhizobia may develop stem nodules on some plant species, create bacteriochlorophyll, and undertake photosynthesis, a unique property of the Bradyrhizobium-legume symbiosis. African wild rice has been shown to contain some photosynthetic bradyrhizobia as endophytes. The significant closeness of 16S rDNA gene sequences is a key complication in determining the taxonomic position and interrelationships of bradyrhizobia. Many strains exhibit 0.1-2.0 percent 16S rDNA sequence divergences. Only B. elkanii and associated strains’ sequences varied by up to 4% from those of other bradyrhizobia. Another problematic element is that these organisms develop extremely slowly, making it difficult to employ traditional phenotypic test protocols (e.g., Biolog, API systems). As a result, numerous bradyrhizobia have been genotypically described more fully. One research has identified at least 11 Bradyrhizobium genospecies, including the described species, utilizing AFLP, DNA-DNA hybridizations, and 16S-23S internal transcribed spacer (ITS) studies. The genera Afipia, Rhodopseudomonas, and Nitrobacter, seem to be closely connected to the bradyrhizobia based on 16S rDNA phylogeny, with B. ekanii having a more peripheral evolutionary position.
According to ITS sequencing data, the photosynthetic bradyrhizobia isolated from Aeschynomene stem-nodules represent a separate group strongly linked to Blastobacter denitrificans. On the other hand, ITS sequencing data reveal that B. elkanii is more closely linked to the bradyrhizobia than the three non-rhizobial taxa. After a thorough investigation of both groups, van Berkumanother plant biologist, advocated renaming Blastobacter denitrificans Bradyrhizobium denitrificans and combining the species with isolates from isolates Aeschynomene indica as Bradyrhizobium denitrificans.

The problem of Rhizobium-Allorhizobium-Agrobacterium based on 16S rDNA sequence data, Azorhizobium and Bradyrhizobium are two different bacteria. Although Sinorhizobium and Mesorhizobium have their clusters, Agrobacterium, Allorhizobium, and Rhizobium are more closely linked. The recent suggestion to merge the genera Agrobacterium and Allorhizobium into Rhizobium has not been universally embraced. For isolates from nodules of Neptunia natans from Senegal, the genus Allorhizobium comprises just one species, Al. undicola. It has a distinct location in the big Agrobacterium-Rhizobium 16S rDNA cluster, with Agrobacterium vitis (96.3 percent 16S rDNA sequence similarity), Rhizobium galegae (95.1 percent), and Rhizobium huautlense (95.3 percent) as its closest neighbors. Due to its dissimilarity to the Rhizobium type species, R. leguminosarum, the muddled taxonomic situation in Agrobacterium and Neptunia isolates can be distinguished phenotypically genotypically from related taxa. It was decided that they should be placed in their genus. This genus may require emendation or revision in a future strategy to rectify the classification and naming of Agrobacterium and Rhizobium species, particularly A. vitis, R. galegae, and R. huautlense. There are now 15 species in the genus Rhizobium, which come from a variety of hosts. There are six recognized species of Agrobacterium, a genus established in 1942that includes bacteria that cause different types of hypertrophies in plants. The first species were mostly described based on phytopathological traits for practical reasons. A. tumefaciens, for example, brings together strains that produce plant tumors; A. radiobacter brings together strains that aren’t dangerous, and A. rhizogenes brings together strains that promote hairy root development A. Rubi is pathogenic on Rubus, A. vitis on the grapevine, and A. larrymoorei on Ficus.
Other nitrogen-fixing symbionts in legumes
Several isolates capable of nitrogen fixation have recently been identified from legume nodules. However, they are phylogenetically outside the typical families of rhizobia in the alpha-Proteobacteria. Methylobacterium, Devosia, Ochrobactrum, and Phyllobacterium are among the new alpha-Proteobacteria lines that include nitrogen-fixing legume symbionts, while Burkholderia, Ralstonia, and Cupriavidus are among the beta-Proteobacteria lines that contain nitrogen-fixing legume symbionts. The following symbiotic, nitrogen-fixing strains have been found in Burkholderia, a genus that encompasses over 20 species of plant pathogens, soil and plant-associated bacteria, and clinical isolates: 2) one strain from Alysicarpus was found to belong to B. cepacia genomovar VI (now B. dolosa, a group previously only found in cystic fibrosis patients; (3) one strain from Machaerium was identified as a new species for which the name B. phymatum has been proposed; and (4) one strain from Aspalathus was identified as a new species for which the name B. Ralstonia taiwanensis was suggested for isolates from Mimosa species in Taiwan, similar to Burkholderia, a genus of plant pathogenic or plant-related, soil and clinical organisms. Devosia neptuniae was suggested for strains from Neptunianatans Indiaand Methylobacteriumnodulans for strains from Crotalaria nodulans identified OchrobactrumLupinus for nodule isolates from Lupinus sp., and Phyllobacterium lupinii for isolates nodulatingTrifolium and Lupinus.

These novel nodulating bacteria are not phylogenetically (16S rDNA) related to rhizobia, but they do have nod genes comparable to rhizobia’s. Nod factors are signal molecules involved in the bacterium-legume communication during nodulation. Some novel nodulating strains outside of the rhizobia have been discovered as a result of investigations of nod gene diversity. These novel nitrogen-fixing legume symbionts most likely acquired the nod genes by lateral gene transfer. The majority of the novel nodulating bacteria are members of genera with at least one plant-associated species, indicating that they are likely to have molecular mechanisms to overcome plant defenses. According to recent findings, more bacteria capable of efficient nodulation will most likely be identified outside of the conventional rhizobia.
